Overview
Hydra, a simple freshwater animal, undergoes continual self-renewal and has robust regenerative capabilities (Figure 1). This is accomplished using the same basic molecular toolkit common to all animals. The aim of our research is to understand the molecular mechanisms that underlie these processes in Hydra, which enables comparative studies animal phylogeny. Hydra is relatively simple with approximately 25 cell types, which allows us to leverage genomic approaches to study development and regeneration at the organismal level.
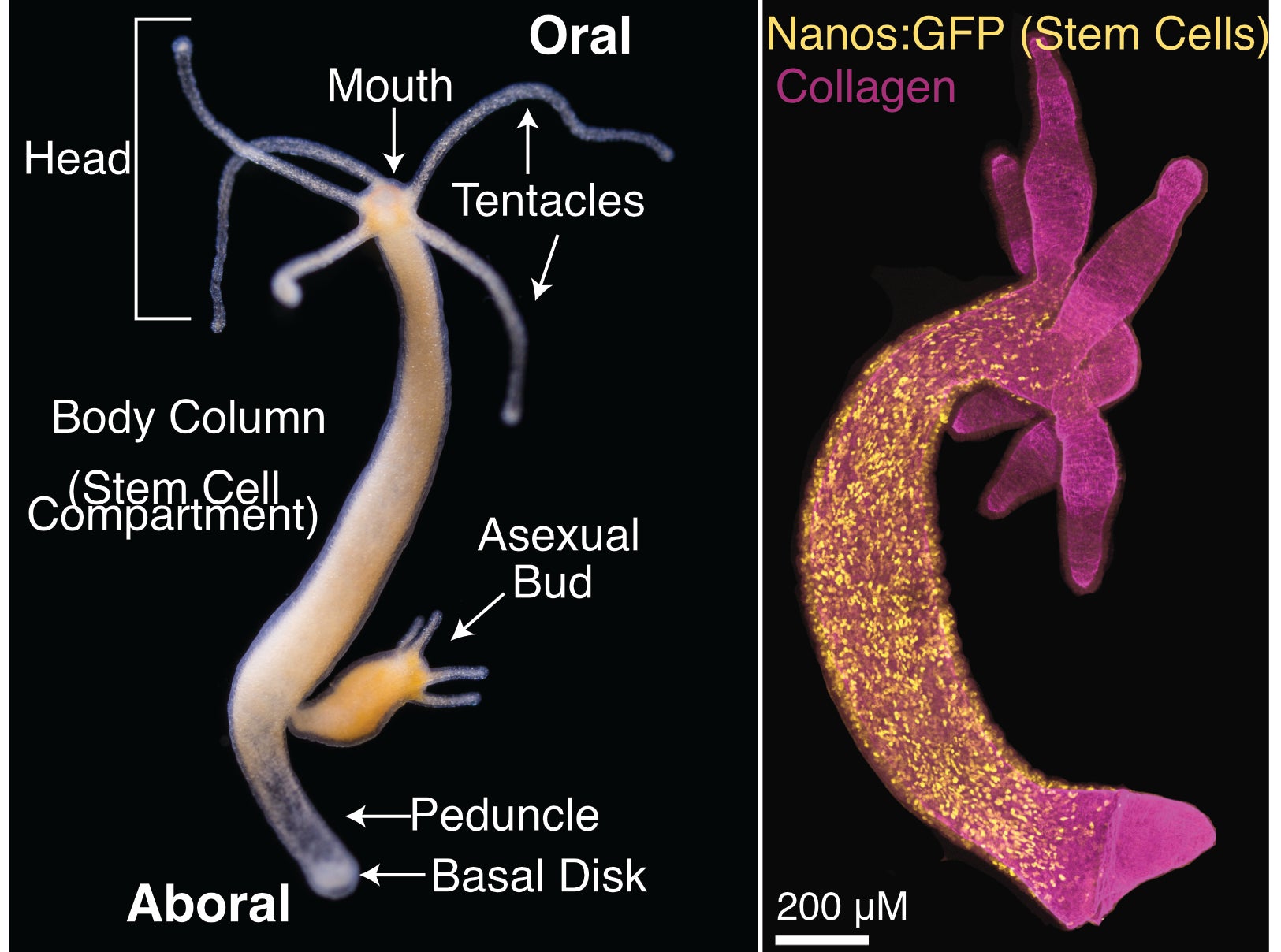
Figure 1 – Hydra vulgaris body plan and stem cell compartment. Hydra vulgaris is organized around a single oral-aboral axis with a head (tentacles and mouth) at the oral end and a peduncle and basal disk at the aboral end (Left Panel, photo credit: Stefan Siebert). A transgenic line expressing GFP in interstitial stem cells (nanos promoter) shows the stem cell compartment is the body column (Right Panel, image credit: Ben Cox).
Gene regulatory networks that that drive homeostatic development at cell type resolution
Hydra vulgaris is composed of three cell lineages (ectoderm, endoderm, and interstitial), each supported by a distinct stem cell population. Continual mitotic divisions of body column epithelial cells replenish the ectodermal and endodermal lineages; as cells translocate toward the oral and aboral ends they terminally differentiate to build the head and foot structures and the oldest cells are shed at the extremities. Cells of the interstitial lineage are closely associated with epithelial cells, and are therefore also continually shed and replaced. Interstitial cell populations (neurons, gland cells, and nematocytes) are maintained by multipotent interstitial stem cells (ISCs). As a result of these tissue movements, differentiation pathways are continuously active.
Single cell RNA sequencing (scRNA-seq) technology is rapidly advancing the field of developmental biology. Collecting thousands of single cell transcriptomes over a developmental time course allows for the ordering of cells into differentiation trajectories by taking advantage of progressive changes in gene expression. This strategy reveals the complete set of genes that underlie cell fate decisions, including rare cell states at key decision branch points, thus giving researchers an unprecedented view of development. We are applying this approach, along with additional genomic approaches to interrogate chromatin state (e.g., ATAC-seq and CUT&Tag) to the adult Hydra. Because all differentiation pathways are active in the adult, this strategy is allowing us to reveal the transcriptional changes that underlie cellular differentiation and build gene regulatory networks (GRNs) to describe the differentiation of all cell types in the homeostatic Hydra. For this work we collaborate with the Farrell lab at the NIH.
Further Reading:
Cazet JF, Siebert S, Morris Little H, Bertemes P, Primack AS, Ladurner P, Achrainer M, Fredriksen MT, Moreland RT, Singh S, Zhang S, Wolfsberg TG, Schnitzler CE, Baxevanis AD, Simakov O, Hobmeyer B, and Juliano CE (2023). A chromosome-scale epigenetic map of the Hydra genome reveals conserved regulators of cell state. Genome Research February 2023. Cover Story. bioRxiv preprint: 10.1101/2022.06.21.496857
Siebert S, Farrell JA, Cazet JF, Abeykoon Y, Primack AS, Schnitzler CE, Juliano CE (2019). Stem cell differentiation trajectories in Hydra resolved at single-cell resolution. Science 365, eaav9314. doi: 10.1126/science.aav9314. bioRxiv preprint: doi:10.1101/460154
Injury-induced activation of developmental pathways during regeneration
Regeneration is activated by injuries that occur at unpredictable times and places, which contrasts the predictability of most developmental processes. Homeostatic maintenance in the adult Hydra vulgaris leads to the predictable replacement of all cells every three weeks. After body part amputation, these differentiation pathways are rapidly activated to replace the missing structure within two days. We are building and comparing the GRNs that direct both homeostatic and regenerative development, with the goal of identifying mechanisms specific to regeneration. Ultimately this will allow us to understand why some animals regenerate well, and others (e.g., humans) do not. We have already uncovered an injury-induced activation of Wnt signaling, a critical developmental pathway, that is specific to the regeneration GRN (Cazet et al., 2021).
Hydra oligactis is closely related to Hydra vulgaris but has reduced regeneration potential. Our data suggest that the regeneration deficiency is caused in part by the inability of the injury response to activate Wnt signaling. H. oligactis therefore provides a test case for inducing regeneration in a non-regenerative context by restoring the connection between injury and developmental pathways, a connection that is likely missing in many nonregenerative organisms.
Further Reading:
Cazet JF, Cho A, Juliano CE (2021) Generic injuries are sufficient to induce ectopic Wnt organizers in Hydra. eLife March 29 2021. doi: 10.7554/eLife.60562. bioRxiv preprint: 10.1101/2020.07.06.189811
Nervous system regeneration at the organismal level
The regenerative powers of H. vulgaris include the ability to rebuild the entire nervous system from adult stem cells. As foundational work towards understanding this remarkable process, we have generated a comprehensive transcriptional map of the uninjured nervous system by sorting and sequencing ~35,000 neurons from a transgenic line that expresses GFP in all neurons and neural progenitors. With these data, we built a comprehensive transcriptomic and spatial map of the nervous system, built trajectories describing the differentiation of all neural subtypes from stem cells, and Built trajectories describing transdifferentiation of neural subtypes (Primack et al., 2023). Our long-term goal is to understand how injury triggers the differentiation and transdifferentiation of appropriate neural subtypes, as well as how these neurons are functionally integrated into neural circuits to restore behavior. To integrate our understanding of nervous system development with nervous system function, we collaborate with the Robinson lab at Rice University.
Further Reading:
Badhiwala KN, Primack AS, Juliano CE, Robinson JT (2021). Multiple nerve rings coordinate Hydra mechanosensory behavior. eLife July 30 2021. doi: 10.7554/eLife.64108. doi: bioRxiv preprint: 10.1101/2020.10.16.343384
Research Presentations:
https://www.youtube.com/watch?v=tdhCPeB3G08&t=2s
Categories
- No categories

 530-752-8882
530-752-8882